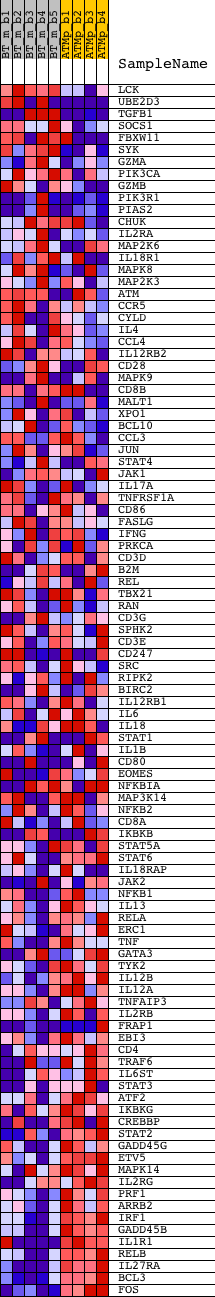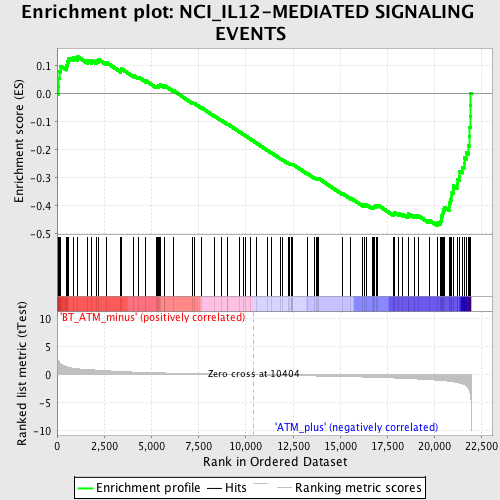
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | NCI_IL12-MEDIATED SIGNALING EVENTS |
| Enrichment Score (ES) | -0.47224554 |
| Normalized Enrichment Score (NES) | -1.8047581 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.24758966 |
| FWER p-Value | 0.693 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | LCK | 1425396_a_at 1439145_at 1439146_s_at 1457917_at | 50 | 2.528 | 0.0273 | No | ||
| 2 | UBE2D3 | 1423112_at 1423113_a_at 1423114_at 1450858_a_at 1450859_s_at 1455479_a_at 1455480_s_at | 77 | 2.421 | 0.0544 | No | ||
| 3 | TGFB1 | 1420653_at 1445360_at | 106 | 2.330 | 0.0803 | No | ||
| 4 | SOCS1 | 1440047_at 1450446_a_at | 198 | 1.904 | 0.0984 | No | ||
| 5 | FBXW11 | 1425461_at 1425462_at 1437468_x_at 1438336_at 1443010_at 1447294_at 1457485_at | 495 | 1.454 | 0.1019 | No | ||
| 6 | SYK | 1418261_at 1418262_at 1425797_a_at 1457239_at | 557 | 1.383 | 0.1152 | No | ||
| 7 | GZMA | 1417898_a_at | 624 | 1.325 | 0.1277 | No | ||
| 8 | PIK3CA | 1423144_at 1429434_at 1429435_x_at 1440054_at 1453134_at 1460326_at | 854 | 1.165 | 0.1308 | No | ||
| 9 | GZMB | 1419060_at | 1103 | 1.058 | 0.1318 | No | ||
| 10 | PIK3R1 | 1425514_at 1425515_at 1438682_at 1444591_at 1451737_at | 1613 | 0.946 | 0.1196 | No | ||
| 11 | PIAS2 | 1419454_x_at 1426456_a_at 1428725_at | 1840 | 0.892 | 0.1196 | No | ||
| 12 | CHUK | 1417091_at 1428210_s_at 1451383_a_at | 2099 | 0.833 | 0.1176 | No | ||
| 13 | IL2RA | 1420691_at 1420692_at | 2200 | 0.808 | 0.1224 | No | ||
| 14 | MAP2K6 | 1426850_a_at 1459292_at | 2630 | 0.715 | 0.1111 | No | ||
| 15 | IL18R1 | 1421628_at | 3371 | 0.583 | 0.0841 | No | ||
| 16 | MAPK8 | 1420931_at 1420932_at 1437045_at 1440856_at 1457936_at | 3414 | 0.576 | 0.0889 | No | ||
| 17 | MAP2K3 | 1425456_a_at 1451714_a_at | 4050 | 0.493 | 0.0656 | No | ||
| 18 | ATM | 1421205_at 1428830_at | 4327 | 0.459 | 0.0583 | No | ||
| 19 | CCR5 | 1422259_a_at 1422260_x_at 1424727_at | 4701 | 0.420 | 0.0461 | No | ||
| 20 | CYLD | 1429617_at 1429618_at 1445213_at | 5240 | 0.367 | 0.0258 | No | ||
| 21 | IL4 | 1449864_at | 5329 | 0.360 | 0.0259 | No | ||
| 22 | CCL4 | 1421578_at | 5373 | 0.355 | 0.0281 | No | ||
| 23 | IL12RB2 | 1421623_at | 5416 | 0.351 | 0.0303 | No | ||
| 24 | CD28 | 1417597_at 1437025_at 1443703_at | 5475 | 0.347 | 0.0317 | No | ||
| 25 | MAPK9 | 1421876_at 1421877_at 1421878_at | 5667 | 0.330 | 0.0268 | No | ||
| 26 | CD8B | 1426170_a_at 1446372_at 1448569_at 1458652_at | 5674 | 0.329 | 0.0304 | No | ||
| 27 | MALT1 | 1445068_at 1456126_at 1456429_at | 6180 | 0.286 | 0.0106 | No | ||
| 28 | XPO1 | 1418442_at 1418443_at 1448070_at | 7177 | 0.210 | -0.0326 | No | ||
| 29 | BCL10 | 1418970_a_at 1418971_x_at 1418972_at 1443524_x_at | 7261 | 0.204 | -0.0340 | No | ||
| 30 | CCL3 | 1419561_at | 7670 | 0.176 | -0.0506 | No | ||
| 31 | JUN | 1417409_at 1448694_at | 8322 | 0.132 | -0.0789 | No | ||
| 32 | STAT4 | 1448713_at | 8711 | 0.107 | -0.0954 | No | ||
| 33 | JAK1 | 1433803_at 1433804_at 1433805_at | 9030 | 0.087 | -0.1089 | No | ||
| 34 | IL17A | 1421672_at | 9036 | 0.087 | -0.1082 | No | ||
| 35 | TNFRSF1A | 1417291_at | 9678 | 0.046 | -0.1370 | No | ||
| 36 | CD86 | 1420404_at 1449858_at | 9864 | 0.034 | -0.1450 | No | ||
| 37 | FASLG | 1418803_a_at 1449235_at | 9986 | 0.027 | -0.1503 | No | ||
| 38 | IFNG | 1425947_at | 10234 | 0.011 | -0.1615 | No | ||
| 39 | PRKCA | 1427562_a_at 1443069_at 1445028_at 1445395_at 1446598_at 1450945_at | 10542 | -0.008 | -0.1754 | No | ||
| 40 | CD3D | 1422828_at | 11144 | -0.046 | -0.2024 | No | ||
| 41 | B2M | 1427511_at 1449289_a_at 1452428_a_at | 11355 | -0.059 | -0.2113 | No | ||
| 42 | REL | 1420710_at | 11816 | -0.088 | -0.2314 | No | ||
| 43 | TBX21 | 1449361_at | 11939 | -0.095 | -0.2358 | No | ||
| 44 | RAN | 1443131_at 1446819_at 1447478_at 1460551_at | 12231 | -0.112 | -0.2479 | No | ||
| 45 | CD3G | 1419178_at | 12305 | -0.117 | -0.2498 | No | ||
| 46 | SPHK2 | 1417431_a_at 1426230_at | 12393 | -0.124 | -0.2524 | No | ||
| 47 | CD3E | 1422105_at 1445748_at | 12436 | -0.126 | -0.2528 | No | ||
| 48 | CD247 | 1420716_at 1426079_at 1426396_at 1428059_at 1428060_at 1438392_at 1452539_a_at | 12463 | -0.128 | -0.2525 | No | ||
| 49 | SRC | 1423240_at 1450918_s_at | 12489 | -0.129 | -0.2521 | No | ||
| 50 | RIPK2 | 1421236_at 1450173_at | 13249 | -0.177 | -0.2848 | No | ||
| 51 | BIRC2 | 1418854_at | 13634 | -0.203 | -0.3000 | No | ||
| 52 | IL12RB1 | 1418166_at | 13738 | -0.210 | -0.3023 | No | ||
| 53 | IL6 | 1450297_at | 13790 | -0.213 | -0.3021 | No | ||
| 54 | IL18 | 1417932_at | 13852 | -0.218 | -0.3024 | No | ||
| 55 | STAT1 | 1420915_at 1440481_at 1450033_a_at 1450034_at | 15109 | -0.307 | -0.3563 | No | ||
| 56 | IL1B | 1449399_a_at | 15545 | -0.340 | -0.3723 | No | ||
| 57 | CD80 | 1427717_at 1432826_a_at 1440592_at 1451950_a_at 1454372_at 1457952_at | 16176 | -0.391 | -0.3965 | No | ||
| 58 | EOMES | 1426001_at 1435172_at | 16289 | -0.401 | -0.3970 | No | ||
| 59 | NFKBIA | 1420088_at 1420089_at 1438157_s_at 1448306_at 1449731_s_at | 16399 | -0.411 | -0.3972 | No | ||
| 60 | MAP3K14 | 1422999_at 1434364_at | 16681 | -0.436 | -0.4050 | No | ||
| 61 | NFKB2 | 1425902_a_at 1429128_x_at | 16780 | -0.446 | -0.4042 | No | ||
| 62 | CD8A | 1440164_x_at 1440811_x_at | 16823 | -0.450 | -0.4009 | No | ||
| 63 | IKBKB | 1426207_at 1426333_a_at 1432275_at 1445141_at 1454184_a_at | 16915 | -0.460 | -0.3997 | No | ||
| 64 | STAT5A | 1421469_a_at 1450259_a_at | 16987 | -0.467 | -0.3975 | No | ||
| 65 | STAT6 | 1421708_a_at 1426353_at | 17837 | -0.561 | -0.4298 | No | ||
| 66 | IL18RAP | 1421291_at 1456545_at | 17842 | -0.562 | -0.4234 | No | ||
| 67 | JAK2 | 1421065_at 1421066_at | 18068 | -0.592 | -0.4268 | No | ||
| 68 | NFKB1 | 1427705_a_at 1442949_at | 18289 | -0.622 | -0.4296 | No | ||
| 69 | IL13 | 1420802_at | 18588 | -0.666 | -0.4355 | No | ||
| 70 | RELA | 1419536_a_at | 18597 | -0.667 | -0.4280 | No | ||
| 71 | ERC1 | 1421501_a_at 1424247_at 1441493_at 1444356_at 1451279_at | 18919 | -0.717 | -0.4343 | No | ||
| 72 | TNF | 1419607_at | 19120 | -0.750 | -0.4347 | No | ||
| 73 | GATA3 | 1448886_at | 19704 | -0.862 | -0.4514 | No | ||
| 74 | TYK2 | 1417306_at | 20161 | -0.971 | -0.4609 | Yes | ||
| 75 | IL12B | 1419530_at 1449497_at | 20298 | -1.004 | -0.4554 | Yes | ||
| 76 | IL12A | 1425454_a_at | 20338 | -1.011 | -0.4454 | Yes | ||
| 77 | TNFAIP3 | 1433699_at 1450829_at | 20369 | -1.017 | -0.4348 | Yes | ||
| 78 | IL2RB | 1417546_at 1448759_at 1459870_x_at | 20434 | -1.032 | -0.4257 | Yes | ||
| 79 | FRAP1 | 1417592_at 1436267_a_at 1446185_at 1459625_at | 20462 | -1.039 | -0.4148 | Yes | ||
| 80 | EBI3 | 1449222_at | 20514 | -1.053 | -0.4048 | Yes | ||
| 81 | CD4 | 1419696_at 1427779_a_at | 20765 | -1.140 | -0.4029 | Yes | ||
| 82 | TRAF6 | 1421376_at 1421377_at 1435350_at 1443288_at 1446940_at 1446977_at | 20770 | -1.143 | -0.3898 | Yes | ||
| 83 | IL6ST | 1421239_at 1437303_at 1452843_at 1460295_s_at | 20813 | -1.163 | -0.3781 | Yes | ||
| 84 | STAT3 | 1424272_at 1426587_a_at 1459961_a_at 1460700_at | 20902 | -1.208 | -0.3680 | Yes | ||
| 85 | ATF2 | 1426582_at 1426583_at 1427559_a_at 1437777_at 1452116_s_at 1459237_at | 20910 | -1.212 | -0.3541 | Yes | ||
| 86 | IKBKG | 1421208_at 1421209_s_at 1435646_at 1435647_at 1450161_at 1454690_at | 20996 | -1.253 | -0.3434 | Yes | ||
| 87 | CREBBP | 1434633_at 1435224_at 1436983_at 1444856_at 1457641_at 1459804_at | 21006 | -1.257 | -0.3291 | Yes | ||
| 88 | STAT2 | 1421911_at 1450403_at | 21181 | -1.359 | -0.3212 | Yes | ||
| 89 | GADD45G | 1453851_a_at | 21192 | -1.365 | -0.3057 | Yes | ||
| 90 | ETV5 | 1420998_at 1428142_at 1450082_s_at | 21313 | -1.468 | -0.2940 | Yes | ||
| 91 | MAPK14 | 1416703_at 1416704_at 1426104_at 1442364_at 1451927_a_at 1459617_at | 21320 | -1.472 | -0.2771 | Yes | ||
| 92 | IL2RG | 1416295_a_at 1416296_at | 21451 | -1.604 | -0.2643 | Yes | ||
| 93 | PRF1 | 1451862_a_at | 21549 | -1.718 | -0.2487 | Yes | ||
| 94 | ARRB2 | 1426239_s_at 1451987_at | 21574 | -1.756 | -0.2292 | Yes | ||
| 95 | IRF1 | 1448436_a_at | 21686 | -2.021 | -0.2107 | Yes | ||
| 96 | GADD45B | 1420197_at 1449773_s_at 1450971_at | 21794 | -2.532 | -0.1860 | Yes | ||
| 97 | IL1R1 | 1448950_at 1459777_at | 21838 | -2.908 | -0.1540 | Yes | ||
| 98 | RELB | 1417856_at | 21846 | -2.947 | -0.1198 | Yes | ||
| 99 | IL27RA | 1449508_at | 21882 | -3.395 | -0.0818 | Yes | ||
| 100 | BCL3 | 1418133_at | 21891 | -3.545 | -0.0407 | Yes | ||
| 101 | FOS | 1423100_at | 21901 | -3.644 | 0.0015 | Yes |